Gadolinium »
PDB 1b9x-4l12 »
1b9y »
Gadolinium in PDB 1b9y: Structural Analysis of Phosducin and Its Phosphorylation-Regulated Interaction with Transducin Beta-Gamma
Protein crystallography data
The structure of Structural Analysis of Phosducin and Its Phosphorylation-Regulated Interaction with Transducin Beta-Gamma, PDB code: 1b9y
was solved by
R.Gaudet,
P.B.Sigler,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 3.00 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 76.470, 89.920, 102.340, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.3 / 26.6 |
Gadolinium Binding Sites:
The binding sites of Gadolinium atom in the Structural Analysis of Phosducin and Its Phosphorylation-Regulated Interaction with Transducin Beta-Gamma
(pdb code 1b9y). This binding sites where shown within
5.0 Angstroms radius around Gadolinium atom.
In total 6 binding sites of Gadolinium where determined in the Structural Analysis of Phosducin and Its Phosphorylation-Regulated Interaction with Transducin Beta-Gamma, PDB code: 1b9y:
Jump to Gadolinium binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Gadolinium where determined in the Structural Analysis of Phosducin and Its Phosphorylation-Regulated Interaction with Transducin Beta-Gamma, PDB code: 1b9y:
Jump to Gadolinium binding site number: 1; 2; 3; 4; 5; 6;
Gadolinium binding site 1 out of 6 in 1b9y
Go back to
Gadolinium binding site 1 out
of 6 in the Structural Analysis of Phosducin and Its Phosphorylation-Regulated Interaction with Transducin Beta-Gamma
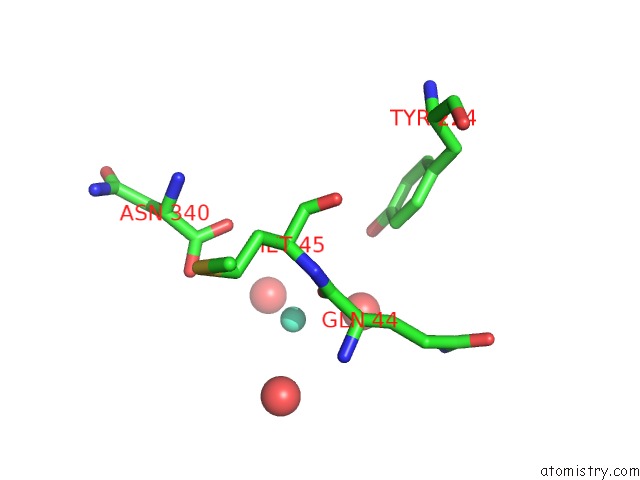
Mono view
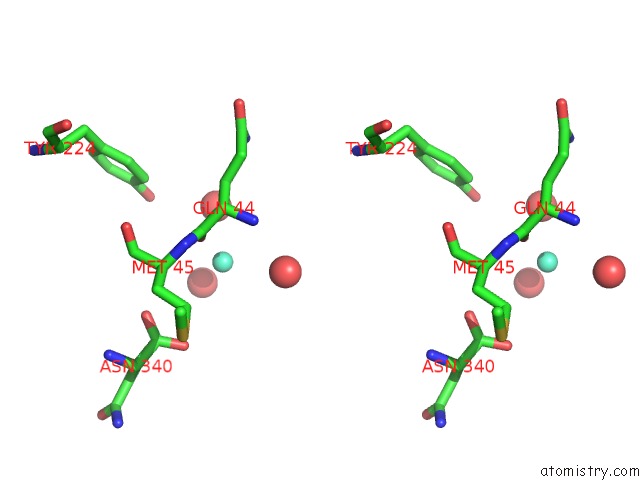
Stereo pair view

Mono view

Stereo pair view
A full contact list of Gadolinium with other atoms in the Gd binding
site number 1 of Structural Analysis of Phosducin and Its Phosphorylation-Regulated Interaction with Transducin Beta-Gamma within 5.0Å range:
|
Gadolinium binding site 2 out of 6 in 1b9y
Go back to
Gadolinium binding site 2 out
of 6 in the Structural Analysis of Phosducin and Its Phosphorylation-Regulated Interaction with Transducin Beta-Gamma

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Gadolinium with other atoms in the Gd binding
site number 2 of Structural Analysis of Phosducin and Its Phosphorylation-Regulated Interaction with Transducin Beta-Gamma within 5.0Å range:
|
Gadolinium binding site 3 out of 6 in 1b9y
Go back to
Gadolinium binding site 3 out
of 6 in the Structural Analysis of Phosducin and Its Phosphorylation-Regulated Interaction with Transducin Beta-Gamma
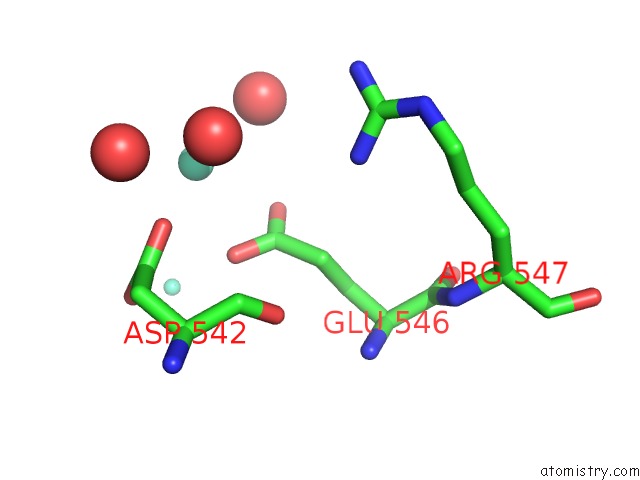
Mono view
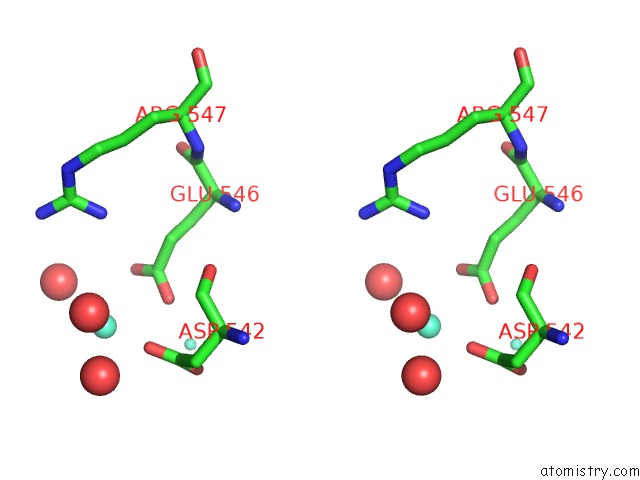
Stereo pair view

Mono view

Stereo pair view
A full contact list of Gadolinium with other atoms in the Gd binding
site number 3 of Structural Analysis of Phosducin and Its Phosphorylation-Regulated Interaction with Transducin Beta-Gamma within 5.0Å range:
|
Gadolinium binding site 4 out of 6 in 1b9y
Go back to
Gadolinium binding site 4 out
of 6 in the Structural Analysis of Phosducin and Its Phosphorylation-Regulated Interaction with Transducin Beta-Gamma
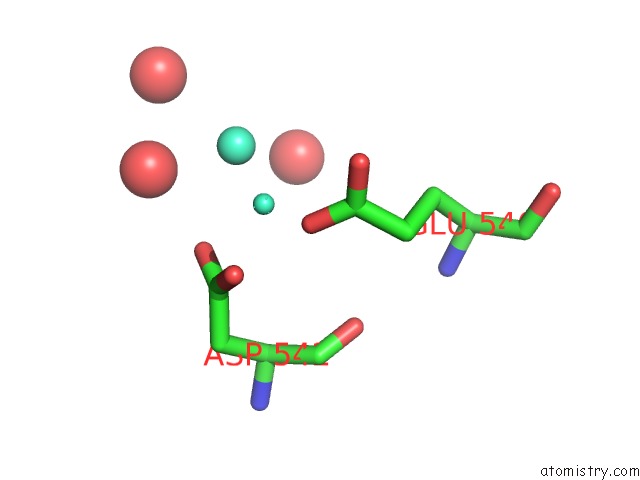
Mono view
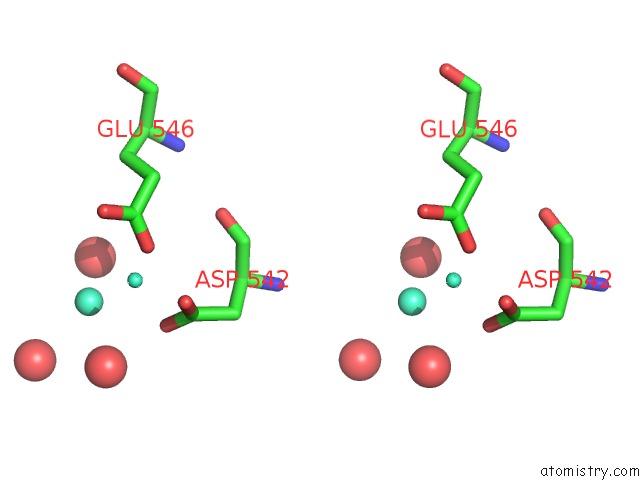
Stereo pair view

Mono view

Stereo pair view
A full contact list of Gadolinium with other atoms in the Gd binding
site number 4 of Structural Analysis of Phosducin and Its Phosphorylation-Regulated Interaction with Transducin Beta-Gamma within 5.0Å range:
|
Gadolinium binding site 5 out of 6 in 1b9y
Go back to
Gadolinium binding site 5 out
of 6 in the Structural Analysis of Phosducin and Its Phosphorylation-Regulated Interaction with Transducin Beta-Gamma
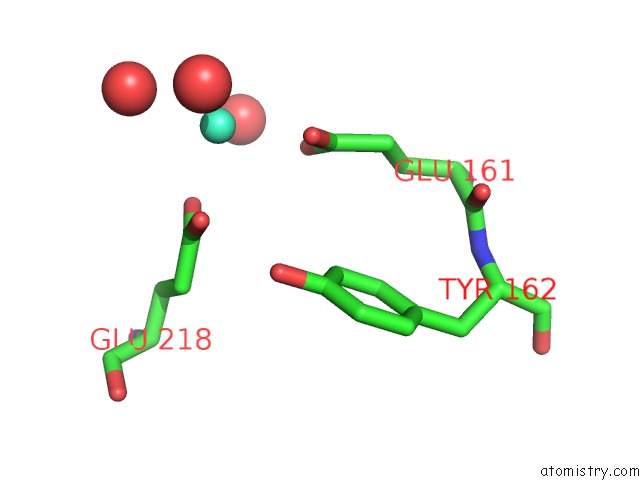
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Gadolinium with other atoms in the Gd binding
site number 5 of Structural Analysis of Phosducin and Its Phosphorylation-Regulated Interaction with Transducin Beta-Gamma within 5.0Å range:
|
Gadolinium binding site 6 out of 6 in 1b9y
Go back to
Gadolinium binding site 6 out
of 6 in the Structural Analysis of Phosducin and Its Phosphorylation-Regulated Interaction with Transducin Beta-Gamma
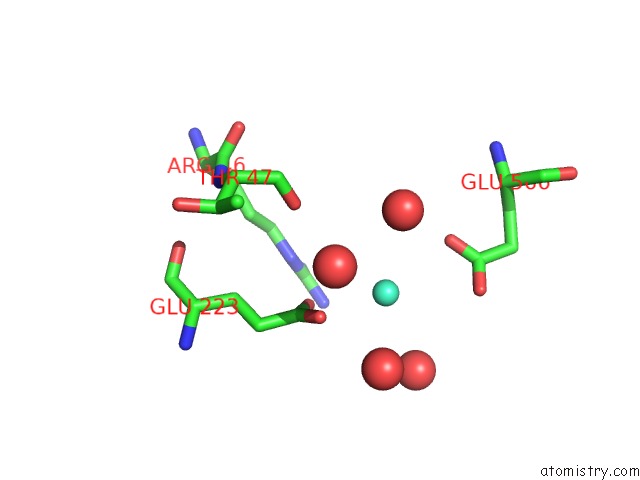
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Gadolinium with other atoms in the Gd binding
site number 6 of Structural Analysis of Phosducin and Its Phosphorylation-Regulated Interaction with Transducin Beta-Gamma within 5.0Å range:
|
Reference:
R.Gaudet,
J.R.Savage,
J.N.Mclaughlin,
B.M.Willardson,
P.B.Sigler.
A Molecular Mechanism For the Phosphorylation-Dependent Regulation of Heterotrimeric G Proteins By Phosducin. Mol.Cell V. 3 649 1999.
ISSN: ISSN 1097-2765
PubMed: 10360181
DOI: 10.1016/S1097-2765(00)80358-5
Page generated: Fri Aug 8 08:24:56 2025
ISSN: ISSN 1097-2765
PubMed: 10360181
DOI: 10.1016/S1097-2765(00)80358-5
Last articles
Zn in 4FUKZn in 4FVD
Zn in 4FVB
Zn in 4FUU
Zn in 4FUP
Zn in 4FUO
Zn in 4FU4
Zn in 4FUN
Zn in 4FUM
Zn in 4FUA