Gadolinium »
PDB 4n5r-6p4z »
5fis »
Gadolinium in PDB 5fis: Exonuclease Domain-Containing 1 (EXD1) in the Gd Bound Conformation
Protein crystallography data
The structure of Exonuclease Domain-Containing 1 (EXD1) in the Gd Bound Conformation, PDB code: 5fis
was solved by
Z.Yang,
K.M.Chen,
R.R.Pandey,
D.Homolka,
M.Reuter,
B.K.Rodino Janeiro,
R.Sachidanandam,
M.O.Fauvarque,
A.A.Mccarthy,
R.S.Pillai,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.01 / 1.60 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 82.860, 135.700, 51.460, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.69 / 18.245 |
Gadolinium Binding Sites:
The binding sites of Gadolinium atom in the Exonuclease Domain-Containing 1 (EXD1) in the Gd Bound Conformation
(pdb code 5fis). This binding sites where shown within
5.0 Angstroms radius around Gadolinium atom.
In total 3 binding sites of Gadolinium where determined in the Exonuclease Domain-Containing 1 (EXD1) in the Gd Bound Conformation, PDB code: 5fis:
Jump to Gadolinium binding site number: 1; 2; 3;
In total 3 binding sites of Gadolinium where determined in the Exonuclease Domain-Containing 1 (EXD1) in the Gd Bound Conformation, PDB code: 5fis:
Jump to Gadolinium binding site number: 1; 2; 3;
Gadolinium binding site 1 out of 3 in 5fis
Go back to
Gadolinium binding site 1 out
of 3 in the Exonuclease Domain-Containing 1 (EXD1) in the Gd Bound Conformation

Mono view
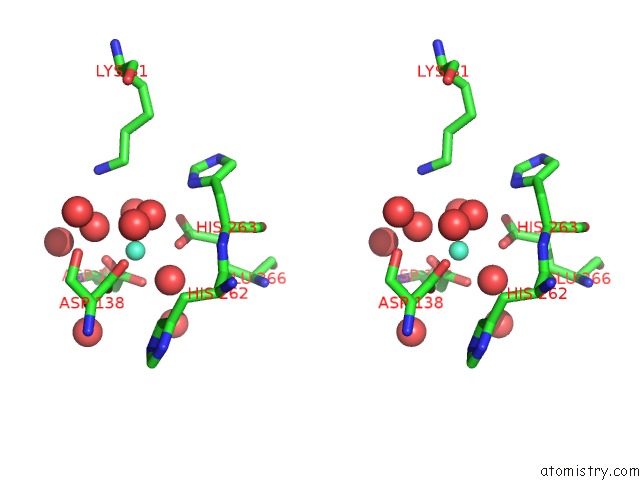
Stereo pair view

Mono view

Stereo pair view
A full contact list of Gadolinium with other atoms in the Gd binding
site number 1 of Exonuclease Domain-Containing 1 (EXD1) in the Gd Bound Conformation within 5.0Å range:
|
Gadolinium binding site 2 out of 3 in 5fis
Go back to
Gadolinium binding site 2 out
of 3 in the Exonuclease Domain-Containing 1 (EXD1) in the Gd Bound Conformation

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Gadolinium with other atoms in the Gd binding
site number 2 of Exonuclease Domain-Containing 1 (EXD1) in the Gd Bound Conformation within 5.0Å range:
|
Gadolinium binding site 3 out of 3 in 5fis
Go back to
Gadolinium binding site 3 out
of 3 in the Exonuclease Domain-Containing 1 (EXD1) in the Gd Bound Conformation
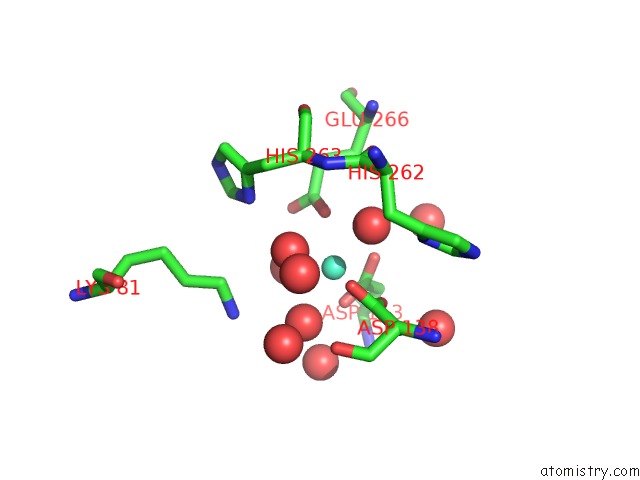
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Gadolinium with other atoms in the Gd binding
site number 3 of Exonuclease Domain-Containing 1 (EXD1) in the Gd Bound Conformation within 5.0Å range:
|
Reference:
Z.Yang,
K.M.Chen,
R.R.Pandey,
D.Homolka,
M.Reuter,
B.K.Rodino Janeiro,
R.Sachidanandam,
M.O.Fauvarque,
A.A.Mccarthy,
R.S.Pillai.
Piwi Slicing and EXD1 Drive Biogenesis of Nuclear Pirnas From Cytosolic Targets of the Mouse Pirna Pathway Mol.Cell V. 61 138 2016.
ISSN: ISSN 1097-2765
PubMed: 26669262
DOI: 10.1016/J.MOLCEL.2015.11.009
Page generated: Sat Aug 10 21:46:28 2024
ISSN: ISSN 1097-2765
PubMed: 26669262
DOI: 10.1016/J.MOLCEL.2015.11.009
Last articles
Zn in 9JPJZn in 9JP7
Zn in 9JPK
Zn in 9JPL
Zn in 9GN6
Zn in 9GN7
Zn in 9GKU
Zn in 9GKW
Zn in 9GKX
Zn in 9GL0