Gadolinium »
PDB 4n5r-6p4z »
4x8i »
Gadolinium in PDB 4x8i: De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase
Enzymatic activity of De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase
All present enzymatic activity of De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase:
3.4.11.15;
3.4.11.15;
Protein crystallography data
The structure of De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase, PDB code: 4x8i
was solved by
M.Colombo,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.06 / 2.50 |
| Space group | I 41 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 203.908, 203.908, 112.443, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.8 / 21.8 |
Other elements in 4x8i:
The structure of De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase also contains other interesting chemical elements:
| Cobalt | (Co) | 3 atoms |
| Zinc | (Zn) | 3 atoms |
| Chlorine | (Cl) | 3 atoms |
Gadolinium Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 11;Binding sites:
The binding sites of Gadolinium atom in the De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase (pdb code 4x8i). This binding sites where shown within 5.0 Angstroms radius around Gadolinium atom.In total 11 binding sites of Gadolinium where determined in the De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase, PDB code: 4x8i:
Jump to Gadolinium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Gadolinium binding site 1 out of 11 in 4x8i
Go back to
Gadolinium binding site 1 out
of 11 in the De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Gadolinium with other atoms in the Gd binding
site number 1 of De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase within 5.0Å range:
|
Gadolinium binding site 2 out of 11 in 4x8i
Go back to
Gadolinium binding site 2 out
of 11 in the De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Gadolinium with other atoms in the Gd binding
site number 2 of De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase within 5.0Å range:
|
Gadolinium binding site 3 out of 11 in 4x8i
Go back to
Gadolinium binding site 3 out
of 11 in the De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase
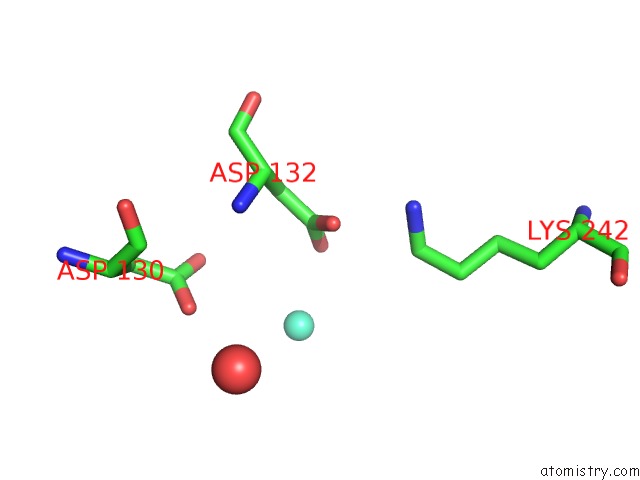
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Gadolinium with other atoms in the Gd binding
site number 3 of De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase within 5.0Å range:
|
Gadolinium binding site 4 out of 11 in 4x8i
Go back to
Gadolinium binding site 4 out
of 11 in the De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Gadolinium with other atoms in the Gd binding
site number 4 of De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase within 5.0Å range:
|
Gadolinium binding site 5 out of 11 in 4x8i
Go back to
Gadolinium binding site 5 out
of 11 in the De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Gadolinium with other atoms in the Gd binding
site number 5 of De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase within 5.0Å range:
|
Gadolinium binding site 6 out of 11 in 4x8i
Go back to
Gadolinium binding site 6 out
of 11 in the De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Gadolinium with other atoms in the Gd binding
site number 6 of De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase within 5.0Å range:
|
Gadolinium binding site 7 out of 11 in 4x8i
Go back to
Gadolinium binding site 7 out
of 11 in the De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase

Mono view
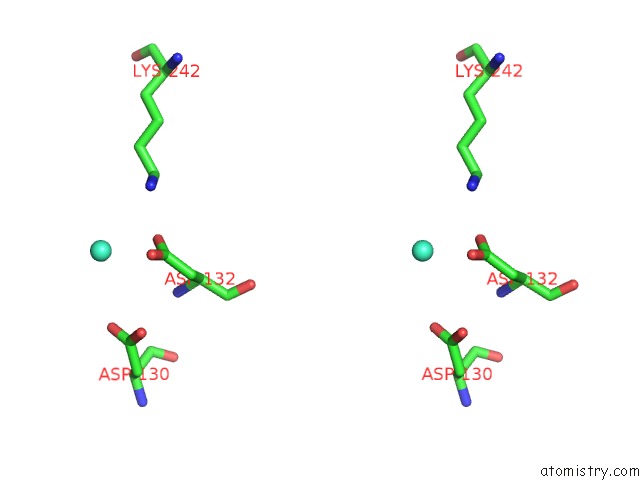
Stereo pair view

Mono view

Stereo pair view
A full contact list of Gadolinium with other atoms in the Gd binding
site number 7 of De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase within 5.0Å range:
|
Gadolinium binding site 8 out of 11 in 4x8i
Go back to
Gadolinium binding site 8 out
of 11 in the De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Gadolinium with other atoms in the Gd binding
site number 8 of De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase within 5.0Å range:
|
Gadolinium binding site 9 out of 11 in 4x8i
Go back to
Gadolinium binding site 9 out
of 11 in the De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Gadolinium with other atoms in the Gd binding
site number 9 of De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase within 5.0Å range:
|
Gadolinium binding site 10 out of 11 in 4x8i
Go back to
Gadolinium binding site 10 out
of 11 in the De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase

Mono view
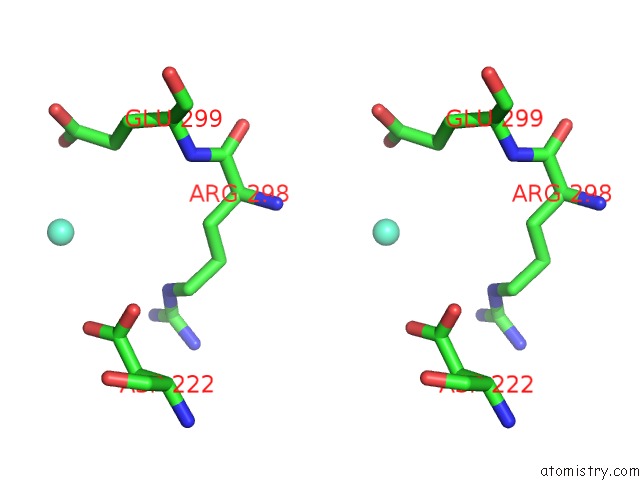
Stereo pair view

Mono view

Stereo pair view
A full contact list of Gadolinium with other atoms in the Gd binding
site number 10 of De Novo Crystal Structure of the Pyrococcus Furiosus TET3 Aminopeptidase within 5.0Å range:
|
Reference:
M.Colombo,
E.Girard,
B.Franzetti.
Tuned By Metals: the Tet Peptidase Activity Is Controlled By 3 Metal Binding Sites. Sci Rep V. 6 20876 2016.
ISSN: ESSN 2045-2322
PubMed: 26853450
DOI: 10.1038/SREP20876
Page generated: Sun Dec 13 19:00:00 2020
ISSN: ESSN 2045-2322
PubMed: 26853450
DOI: 10.1038/SREP20876
Last articles
Zn in 8WB0Zn in 8WAX
Zn in 8WAU
Zn in 8WAZ
Zn in 8WAY
Zn in 8WAV
Zn in 8WAW
Zn in 8WAT
Zn in 8W7M
Zn in 8WD3